The Algorithm
The algorithm builds the entire candidate-solution space (size is \( \sum_{r=1}^n {n \choose r} = {n \choose 1} + {n \choose 2} + \cdot \cdot \cdot + {n \choose n}= 2^n-1 \), where \(n\) is the number of starting features) and uses dynamic programming to efficiently search for optimal solutions from it.
It turns out the candidate-solution space does indeed have the two qualities required in order to leverage Dynamic Programming:
- Optimal Substructure
- Overlapping Sub-problems
The solution space is, therefore, built from the bottom up. The idea is to utilize these properties in order to avoid traversing every combination of \({n \choose k}\) features, where \(n\) is the total number of continuous features that we start with and \(k\) varies from \(1\) to \(n\).
Thus, feature-combinations occurring deeper in the tree will be consisered non-optimal (and thereby discarded) if they do not include optimal feature combinations occurring earlier in the tree. Therefore, by using a Dynamic Programming approach, we avoid needlessly recomputing and re-testing optimal sub-problems that have already been encountered.
Cross-validation over 5 k-folds is used with a scoring method to select the model (built on training data) that produces the least RMSE and the difference between that RMSE vs. the RMSE computed on the testing data, with Condition Number \(\le 100\) (in order to minimize colinearity). This basis is taken directly from statsmodels Github source code for the OLS fit results summary method but I restrict it even further (statsmodels defines non-colinearity by virtue of this value being less than 1000). (“statsmodels.regression.linear_model — statsmodels v0.11.0rc1 (+56): RegressionResults.summary() method source code”, 2019)
In this way, we minimize residuals and thereby select the most predictive model, based on the “best” (minimized \(RMSE\)) non-colinear feature-combination subset from the starting set of all features.
The procedure for this is summarized below in pseudo-code:
COMMENT: let \( ofs \) denote the list of optimal feature subsets (this holds the table of optimal sub-problems)
set \( ofs := \) new list
for \( k := 1 \) to \( n \) (where \( n := |{starting\ features}| \))
COMMENT: let \( fs \) denote the list of all feature subsets where each \( fs[k] \) holds all lists of \( k \) features from \( {n \choose k}=\frac{n!}{k! \cdot (n-k)!} \) (from \( n \) starting features)
set \( fs := \) build all subsets of \( k \) features from \( {n \choose k} \)
set \(depth := k - 1\)
COMMENT: let \( kfs \) denote a subset of \( fs \) with \( k \) features
for each \( kfs \) in \( fs \)
COMMENT: let \( cpd \) denote the closest prior depth
set \( cpd \) \( := min(len(ofs)-1, depth-1) \)
COMMENT: qualify that current \( kfs \) is built from the last optimal sub-problem already computed - if not, then discard it
if \( depth > 0 \) and \( cpd \ge 0 \) then
COMMENT: let \( lofc \) denote the last optimal feature combination (from a prior depth)
set \( lofc := ofs[cpd] \)
if \( lofc \) not in \( kfs \) then
COMMENT: discard this \( kfs \) since it does not contain the last optimal feature combination and then loop to the next \( kfs \)
continue
COMMENT: otherwise this \( kfs \) contains \( lofc \) (or \( depth==0 \) and this \( kfs \) is embryonic)
set \( kfolds := \) build 5-kfolds based on \( kfs \)
for each \( fold \) in \( kfolds \) {
split data set into \( partition_{test} \) and \( partition_{train} \)
COMMENT: let \( lrm \) denote the linear regression model built from training data from this \( fold \)
set \( lrm := \) build linear regression from \( partition_{train} \)
set \( target_{train, predicted} := \) compute predictions with \( lrm \) from \( partition_{train} \)
set \( target_{test, predicted} := \) compute predictions with \( lrm \) from \( partition_{test} \)
set \( RMSE_{train} := \) compute Root Mean Squared Error between \( target_{train, actual} \) and \( target_{train, predicted} \) (i.e. - \( RMSE \) of residuals of \( partition_{train} \))
set \( RMSE_{test} := \) compute Root Mean Squared Error between \( target_{test, actual} \) and \( target_{test, predicted} \) (i.e. - \( RMSE \) of residuals of \( partition_{test} \))
COMMENT: let \( sl_{fold} \) denote the list of tracked scores for each \( fold \)
append \( (RMSE_{train}, RMSE_{test}) \) to \( sl_{fold} \)
set \( sl_{fold, RMSE_{train}} := \) extract all \( RMSE_{train} \) from \( sl_{fold} \)
set \( RMSE := \frac{\sum RMSE_{train}}{size(sl_{fold, RMSE_{train}})} \)
set \( sl_{fold, RMSE_{test}} := \) extract all \( RMSE_{test} \) from \( sl_{fold} \)
set \( \Delta RMSE := \frac{\sum |RMSE_{train} - RMSE_{train}|}{size(sl_{fold, RMSE_{train}})} \)
if \( RMSE_{best} \) is null then
set \( RMSE_{best} := RMSE \)
set \( \Delta RMSE_{best} := \Delta RMSE \)
else
if \( RMSE < RMSE_{best} \) AND \( lrm.condition\_number \le 100 \) then
set \(RMSE_{best} := RMSE\)
set \( \Delta RMSE_{best} := \Delta RMSE \)
set \( ofs[depth] := kfs \)
This results in cross-validation selecting the best non-colinear feature-combination subset for each \( k \), from \( n \) starting features, that predicts the outcome, price, with the greatest accuracy (lowest \( \Delta RMSE \)).
The total number of all possible combinations the algorithm will select from is \( \sum_{r=1}^n {n \choose r} = {n \choose 1} + {n \choose 2} + \cdot \cdot \cdot + {n \choose n}= 2^n-1 \), but it avoids traversing that entire space by leveraging dynamic programming.
That number can grow quite large rather quickly.
For instance, starting with \( n=18 \) features, we have \( \sum_{r=1}^{18} {18 \choose r} = 2^{18}-1 = 262143 \) possible combinations!
But, because it uses the dynamic programming approach (vs. brute force), this algorithm is nevertheless fast, all things considered.
Source Code
The full source code is written in Python, specifically for Jupyter Notebooks, and was uploaded to this github repository. But it can be adapted for use outside of Juptyer Notebooks as you wish.
The essence is captured in the following function, a snippet from full source code:
def cv_selection_dp(
X
, y
, folds=5
, reverse=False
, smargs=None):
cols = ['n_features', 'features', condition_no, rsquared, adjusted_rsquared, pvals, rmse, delta_rmse]
scores_df = pd.DataFrame(columns=cols)
target_cond_no = None
if smargs is not None:
target_cond_no = smargs['cond_no']
if target_cond_no is None:
target_cond_no = 1000 #default definition of "non-colinearity" used by statsmodels - see https://www.statsmodels.org/dev/_modules/statsmodels/regression/linear_model.html#RegressionResults.summary
cv_feat_combo_map, _ = cv_build_feature_combinations(X, reverse=reverse)
if cv_feat_combo_map is None:
return
base_feature_set = list(X.columns)
n = len(base_feature_set)
best_feat_combo = []
best_score = None
best_feat_combos = []
for _, list_of_feat_combos in cv_feat_combo_map.items():
n_choose_k = len(list_of_feat_combos)
k = len(list_of_feat_combos[0])
depth = k-1
s_n_choose_k = "{} \\choose {}"
display(
HTML(
"<p><br><br>Cross-validating ${}={}$ combinations of {} features (out of {}) over {} folds using score <b>{}</b> and target cond. no = {}...".format(
"{" + s_n_choose_k.format(n, k) + "}"
, n_choose_k
, k
, n
, folds
, condition_no_and_pvals_and_rsq_and_adjrsq_and_rmse_and_delta_rmse
, target_cond_no
)
)
)
n_discarded = 0
n_met_constraints = 0
for feat_combo in list_of_feat_combos:
feat_combo = list(feat_combo)
closest_prior_depth = min(len(best_feat_combos)-1, depth-1)
if depth > 0 and closest_prior_depth >= 0:
last_best_feat_combo = best_feat_combos[closest_prior_depth]
last_best_feat_combo_in_current_feat_combo = set(last_best_feat_combo).issubset(set(feat_combo))
if last_best_feat_combo_in_current_feat_combo:
#print("depth is {}, best_feat_combos[last_saved_depth]: {}".format(depth, best_feat_combos[last_saved_depth]))
#print("feat_combo: {}".format(feat_combo))
#print("best_feat_combos[last_saved_depth] in feat_combo: {}".format(last_best_feat_combo_in_current_feat_combo))
pass
else:
n_discarded += 1
#display(HTML("DISCARDED feature-combo {} since it is not based on last best feature-combo {}; discarded so far: {}".format(feat_combo, last_best_feat_combo, n_discarded)))
continue
_, _, _, _, score = cv_score(
X
, y
, feat_combo
, folds
, condition_no_and_pvals_and_rsq_and_adjrsq_and_rmse_and_delta_rmse
)
#now determine if this score is best
is_in_conf_interval = False not in [True if pval >= 0.0 and pval <= 0.05 else False for pval in score[3]]
is_non_colinear = score[0] <= target_cond_no
if is_non_colinear and is_in_conf_interval and (best_score is None or (score[1] > best_score[1] and score[2] > best_score[2])):
n_met_constraints += 1
best_score = score
best_feat_combo = feat_combo
if len(best_feat_combos) < k:
best_feat_combos.append(feat_combo)
else:
best_feat_combos[depth] = feat_combo
print(
"new best {} score: {}, from feature-set combo: {}".format(
condition_no_and_pvals_and_rsq_and_adjrsq_and_rmse_and_delta_rmse
, best_score
, best_feat_combo
)
)
data = [
{
'n_features': len(feat_combo)
, 'features': feat_combo
, condition_no: score[0]
, rsquared: score[1]
, adjusted_rsquared: score[2]
, pvals: score[3]
, rmse: score[4]
, delta_rmse: score[5]
}
]
mask = scores_df['n_features']==k
if len(scores_df.loc[mask]) == 0:
scores_df = scores_df.append(data, ignore_index=True, sort=False)
else:
keys = list(data[0].keys())
replacement_vals = list(data[0].values())
scores_df.loc[mask, keys] = [replacement_vals]
if n_discarded > 0:
display(
HTML(
"<p>DISCARDED {} {}-feature combinations that were not based on prior optimal feature-combo {}".format(
n_discarded
, k
, last_best_feat_combo
)
)
)
if n_met_constraints > 0:
display(
HTML(
"<p>cv_selection chose the best of {} {}-feature combinations that met the constraints (out of {} considered)".format(
n_met_constraints
, k
, n_choose_k - n_discarded
)
)
)
if n_choose_k - n_discarded - n_met_constraints > 0:
display(
HTML(
"<p>{} {}-feature combinations (out of {} considered) failed to meet the constraints<p><br><br>".format(
n_choose_k - n_discarded - n_met_constraints
, k
, n_choose_k - n_discarded
)
)
)
display(HTML("<h2>Table of cv_selected Optimized Feature Combinations</h2>"))
print_df(scores_df)
display(HTML("<h4>cv_selected best {} = {}</h4>".format(condition_no_and_pvals_and_rsq_and_adjrsq_and_rmse_and_delta_rmse, best_score)))
display(
HTML(
"<h4>cv_selected best feature-set combo ({} of {} features) {} based on {} scoring method with target cond. no. {}</h4>".format(
len(best_feat_combo)
, len(base_feature_set)
, best_feat_combo
, condition_no_and_pvals_and_rsq_and_adjrsq_and_rmse_and_delta_rmse
, target_cond_no
)
)
)
display(HTML("<h4>starting feature-set:{}</h4>".format(base_feature_set)))
to_drop = list(set(base_feature_set).difference(set(best_feat_combo)))
display(HTML("<h4>cv_selection suggests dropping {}.</h4>".format(to_drop if len(to_drop)>0 else "<i>no features</i> from {}".format(base_feature_set))))
return (scores_df, best_feat_combo, best_score, to_drop)
Example
I have written a sample Jupyter Notebook that utilizes the API and uploaded it to the same repository in which the source code is found.
Here are a few snippets from that notebook.
Snippet 1:

Snippet 2:
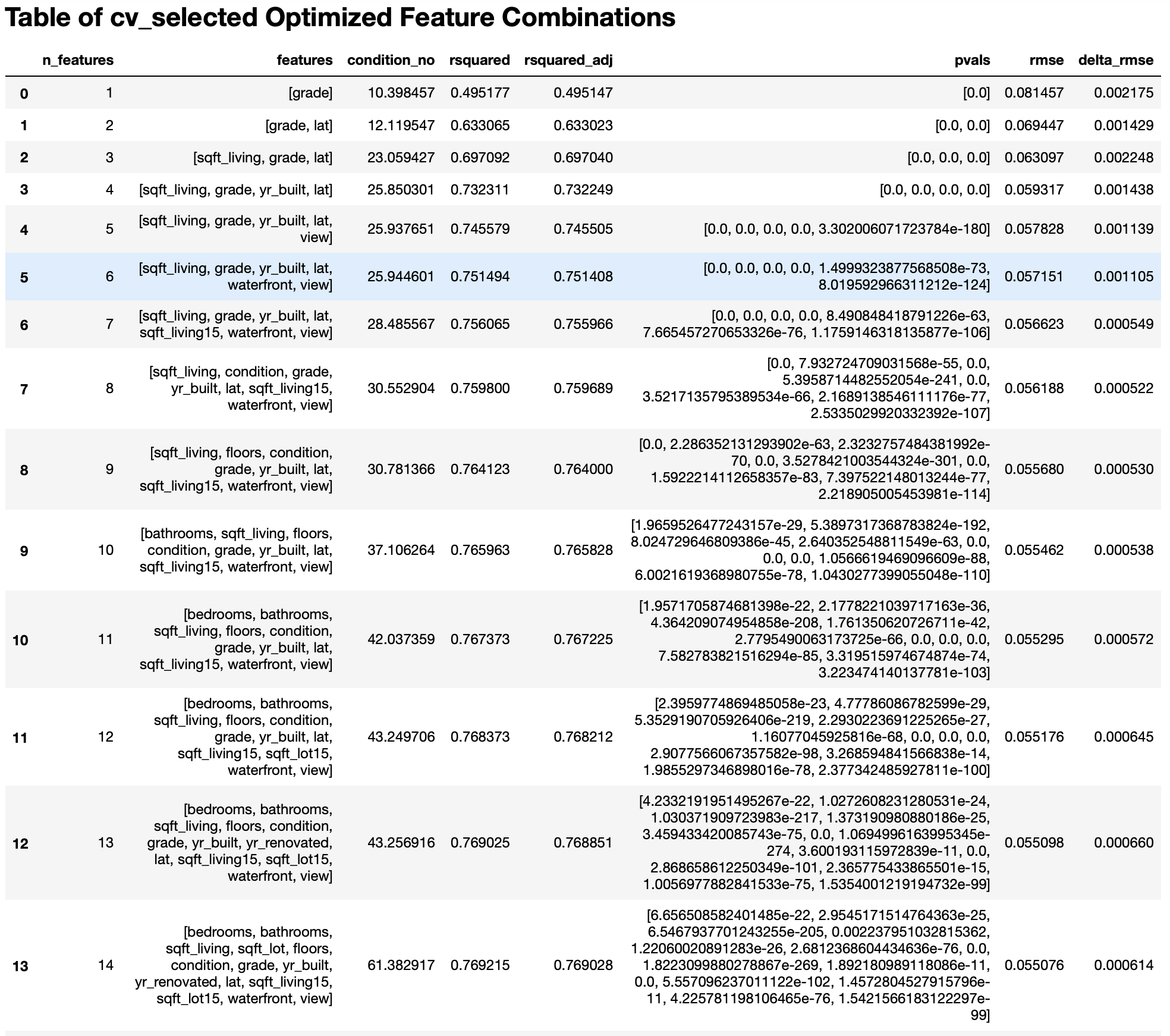
Snippet 3:

The snippets above are just that: snippets and are thus incomplete. To see the full results, please refer to the example Jupyter notebook.
References
Gavis-Hughson, S. (2019). The Ultimate Guide to Dynamic Programming - Simple Programmer. Retrieved from https://simpleprogrammer.com/guide-dynamic-programming/
statsmodels.regression.linear_model — statsmodels v0.11.0rc1 (+56): RegressionResults.summary() method source code. (2019). Retrieved from https://www.statsmodels.org/dev/_modules/statsmodels/regression/linear_model.html#RegressionResults.summary
Rabbit, B. (2018). Revisions to Is there a math nCr function in python? [duplicate]. Retrieved from https://stackoverflow.com/posts/4941932/revisions